Products
FIB-SEM
Nanomanipulators
OmniProbeOmniProbe CryoSoftware
AZtec3DAZtecFeatureAZtec LayerProbeTEM
Hardware
EDSUltim MaxXploreImaging
TEM CamerasSoftware
AZtecTEM
14th April 2022 | Author: Richard McLaughlin
AZtec EDS and EBSD are powerful systems that can generate large amounts of data. AZtec supplies tools to analyse and present data in numerous ways, but some users may have a unique application requiring data manipulation that AZtec does not provide. In these situations, the user must export the raw data and then process it for themselves. Fortunately, AZtec can export data in various formats, from comma separated variable files, tab separated variable files, EMSA, and Ripple, to name just a few.
Starting in AZtec version 6.0, a new way to export has been introduced, the H5OINA file format. Using the Hierarchical Data Format 5 file format library, H5OINA is optimised for I/O speed when accessing large datasets. Other benefits include an open specification, so no proprietary road blocks to accessing the data, and bindings to the most popular programming languages.
In this blog, we are going to help the new user set up their tools to extract data from the H5OINA file using one of the more popular programming languages among scientists, Python.
Obviously, we need Python, and if it’s not already installed, go to www.python.org to download the latest version and install it on to the computer you plan to use for data processing. The next item we need to download is the HDF5 library that allows easy access to the H5OINA file using only a few programming statements. If you are using Fortran, C, or C++, you will need to go to www.hdf5group.org and download the library. It’s not necessary, but you can also download the official HDF Viewer from the same website. It’s a small application that allows you to open and view the data structure of the file and the included datatypes. It can be a great aid in understanding how and what to extract.
With Python, downloading the library is as simple as typing, “pip install h5py”. Below, I have opened the command Prompt on a Windows 10 computer, with Python installed, and typed in the command. If you are connected to the internet, the HDF5 libraries will be automatically downloaded and installed within seconds.
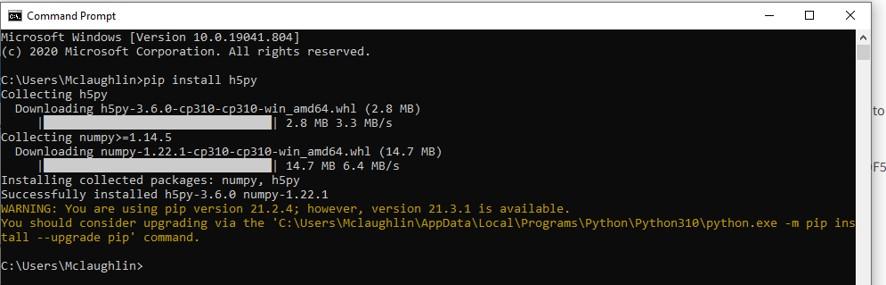
Python is well known for its ease of use and simplicity. All you need is Windows Notepad or some other text editor to write your script. Once written, you can run the script by typing “python myscript.py”. Replace “myscript.py” with the name of the script you have created.
Although it’s not necessary, you can download and use Visual Studio Code as an editor for your scripts. It provides useful tools and a nice interface for creating your scripts. It can be downloaded here. Make sure to add the Python language Extension!
For both examples, we are going to be using the same H5OINA file containing data from an AZtec Feature analysis of steel inclusions.
We will start with a simple example where the goal is to extract one ‘raw’ spectrum and save it to a text file. The file containing the data, “Steel.h5oina”, is to be opened for reading, and a file “spectrum.txt” is to be created and opened for writing. The spectrum associated with Feature 1000 will be read and the counts in each channel will be written to the text file with the count values separated by a comma. The script that will do this is presented below on the left. On the right, is the view of data structure in our Steel.h5oina file as shown in the HDF Viewer.

The most important lines are 7 through 9. On line 7, we assign the data object (one dimensional array) of interest to variable “spectrum_Data”, and on line 9 and 10, we iterate through the array reading each value and then writing it to our text file.
Well, that seems like a lot of work to extract one spectrum, but what if you wanted to extract raw data from thousands of spectra? The next example will do exactly that.
The following script extracts all spectra data from the Steel.h5oina file and writes the data to a text file called “spectra.txt”. The challenge here is to figure out the Feature IDs of all the inclusions, then use that information to create the path to each stored spectrum. Fortunately, there is an “Index” dataset that holds all the Feature IDs. We can read the index list into a variable “feature_Index”, then read each Feature ID and add this to our path. For each spectrum, we just need to repeat what we did in the Example 1. Below on the left, is the Script, and on the right, is the data structure shown in HDF Viewer.

The new H5oina file format is a response to the changing demands of users and their need to have an easy and quick way to access their large datasets now and into the future. As shown in this blog, in a few minutes, anyone can set up their computer to access and process raw X-ray maps, spectra, or images exactly the way they want and without any limitations.
We hope this shows how export to the H5OINA data format makes it easy to access and process data in ways that are bespoke to your needs. If you want to learn about the more general export options in AZtec then why not try these recent blog posts.
We send out monthly newsletters keeping you up to date with our latest developments such as webinars, new application notes and product updates.