There are many challenges with imaging biological specimens using electron microscopy and most of them also apply to energy dispersive X-ray spectrometry (EDS). The interior of an electron microscope is a hostile environment for life; the vacuum and high energy electron beam are both destructive for the soft materials in most biological samples. As biologists, we want to capture data that is as close to the living state as possible, which means careful preservation of our specimens to avoid changing their structure and contents. This is particularly important when we need to preserve elemental composition.
The importance of fixation
Electron microscopy needs some degree of vacuum inside the microscope, which in turn requires that all samples are dehydrated and dried or frozen to avoid water evaporation and distortion of the sample upon exposure. Dehydration causes the clumping together of structures and molecules within cells and tissues, and their relative positioning must be secured prior to this via a process called fixation. Fixation halts all metabolic functions (kills the sample) and ensures that everything stays in place. It can be done using chemicals that form permanent bonds or by freezing the sample very fast to avoid the formation of ice-crystals.
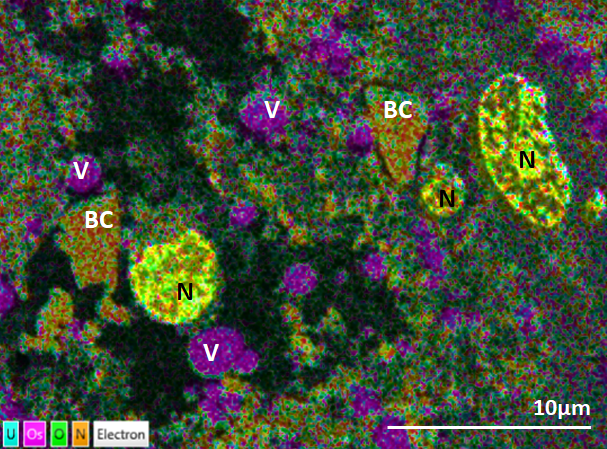
This 100 nm section of liver tissue has been imaged uncoated using an accelerating voltage of 4kV. The liver sample was fixed, stained with osmium tetroxide and uranyl acetate and embedded in Epon 812 resin. Even with only 4 elements selected in this image, clear differentiation between cell nuclei (N), vesicles (V) and blood cells (BC) can be seen. Data was captured using the Ultim Extreme detector.
The need for contrast
In addition to fixation and dehydration, it is important to be able to see the sample and details of its structure, which involves creating some contrast. Contrast is usually achieved by applying a stain that has a high enough density to be visualised with electrons, in the same way that coloured stains are applied to see samples with a light microscope. Stains allow us to image structures such as membranes that we would not normally be able to see. While staining is not necessary for EDS, it is helpful to have an idea of where in the cell or tissue elements detected by EDS are based.
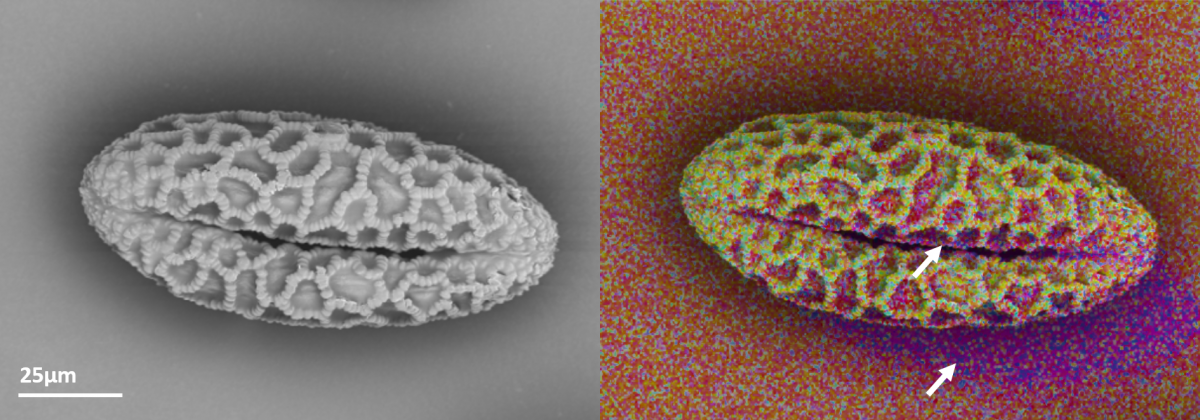
A backscattered electron image (left) and EDS map (right) of a lily pollen grain. There is an artificial shadow (arrows) in the EDS data caused by the detector sitting on one side of the microscope chamber and some x-rays being absorbed by the sample. Data was collected using an Ultim Extreme.
Avoiding the shadows
Biological samples are three-dimensional by their nature, and this complicates EDS data collection. EDS detectors typically sit on one side of the microscope chamber. Uneven or curved samples create an artificial shadowing where X-rays emitted away from the detector are likely to be absorbed by the sample, leading to lower count rates compared to the side facing the detector. This will result in artefacts in elemental maps of the sample, especially for elements with low energy X-rays (particularly relevant for the light elements common in biology).
A flat surface is the best option for good EDS analysis. Some samples are naturally flat, for example, the surface of a leaf, and can be imaged and analysed directly. Thicker or more uneven samples can be split or fractured to expose a flatter surface. This can be difficult and is often unpredictable but is a fast and easy way to achieve a result.
Alternatively, specimens can be embedded in resin and trimmed with a knife so that the area of interest is exposed on a flat plane. The specimen block can be mounted, whole, onto an SEM stub and imaged. Instead of the block, fine slices of the resin-embedded sample can be cut using a microtome and collected onto a substrate for imaging in the SEM. Both options offer a precise way to prepare a flat surface and have the added benefit of enabling us to also collect samples for light microscopy or transmission electron microscopy from the same specimen.
Improved conductivity reduces distortions
Biological samples are non-conductive, and this includes resin-embedded samples. A normal biological sample can’t conduct away the negative charge from an electron beam, and it will build up in the sample, often called charging. This results in distorted images and the sample appearing to move or drift under the beam. EDS data collection can take some time and the sample must be stable during data collection. Conductivity can be improved by using silver paint or conductive epoxy when mounting the sample onto the stub, by adding heavy metal stain during specimen preparation and by coating the sample and stub with carbon or a metal coating. Carbon is the standard coating material for biological samples and EDS.
Low accelerating voltages avoid damage
Finally, even when a well-prepared sample is conductive and stable in the microscope it is still fragile. High beam currents and accelerating voltages can easily damage a sample before data can be collected. It is important to consider microscope conditions and the type of EDS detector being used. Most biological samples will benefit from using a low accelerating voltage to avoid damage and reduce the amount of specimen-beam interaction. Oxford Instruments’ windowless Ultim Extreme detector is the logical choice for most biological applications, maximising spatial resolution and sensitivity in order to achieve results with sensitive samples such as biological material.
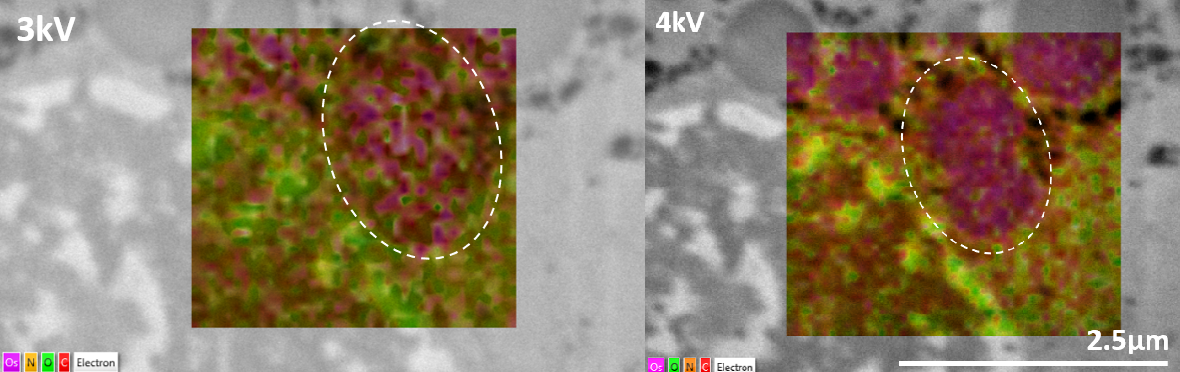
Microscope operating conditions not only affect the sample but also the data produced. This is particularly important with low accelerating voltages and the X-ray lines of particular elements may not be excited. Osmium, a common stain for biological samples in the form of osmium tetroxide, is not detected within liver vesicles at an accelerating voltage of 3 kV (left, dashed region) but can clearly be observed once it is increased to 4 kV (right image). Data was captured using the Ultim Extreme.
If you want to find out more about our solutions for life science applications, watch our webinar or get in touch.




